Researchers From NVIDIA & Vanderbilt University Propose ‘Swin UNETR’: A Novel Architecture for Semantic Segmentation of Brain Tumors Using Multi-Modal MRI Images
The human brain is affected by about 120 different forms of brain tumors. AI-based intervention for tumor identification and surgical pre-assessment is on the verge of becoming a necessity rather than a luxury as we approach the era of artificial intelligence (AI) in healthcare. Brain tumors can be characterized in depth using techniques like volumetric analysis to track their growth and aid in pre-surgical planning.
The characterization of defined tumors can be directly used for the prognosis of life expectancy, in addition to surgical uses. The segmentation of brain tumors is at the forefront of all such applications. Primary and secondary tumors are the two forms of brain tumors. Primary brain cancers arise from brain cells, whereas secondary tumors spread from other organs to the brain.
Gliomas, which emerge from brain glial cells and are divided into low-grade (LGG) and high-grade (HGG) subtypes, are the most frequent primary brain tumors. High-grade gliomas are a form of malignant brain tumor that proliferates and requires surgery and radiotherapy to treat. They have a terrible prognosis.
Magnetic Resonance Imaging (MRI) is a proven diagnostic technology that is used to monitor and plan surgery for brain tumor analysis. To emphasize diverse tissue features and areas of tumor dissemination, many complementary 3D MRI modalities, such as T1, T1 with contrast agent (T1c), T2, and Fluid-attenuated Inversion Recovery (FLAIR), are typically required.
Furthermore, automated medical image segmentation techniques have emerged as a popular method for delineating brain tumors that is both accurate and repeatable. Deep learning-based brain tumor segmentation approaches have recently obtained best-in-class results in a variety of benchmarks.
Convolutional Neural Networks (CNNs) have significant feature extraction capabilities, which have enabled these advancements. However, CNN-based approaches’ small kernel size limits their ability to learn long-range relationships, which are necessary for accurate tumor segmentation in a variety of forms and sizes. Although various attempts have been made to overcome this constraint by extending the effective receptive field of convolutional kernels, the effective receptive field remains confined to local regions.
Vision Transformers (ViTs) have shown state-of-the-art performance on a variety of benchmarks in computer vision. In ViT-based models, the self-attention module enables long-range modeling information through pairwise interaction between token embeddings, resulting in more effective local and global contextual representations. Furthermore, ViTs have demonstrated successful learning of pretext tasks for self-supervised pre-training in a variety of applications.
UNETR is the first method in medical image analysis to use a ViT as an encoder rather than a CNN-based feature extractor. Other approaches have sought to use ViTs as a stand-alone component in architectures that are otherwise made up of CNN-based components. UNETR, on the other hand, has demonstrated superior accuracy and efficiency in a variety of medical picture segmentation tasks.
Swin transformers, a hierarchical vision transformer that computes self-attention in an efficient shifting window partitioning approach, have recently been proposed. Swin transformers are thus appropriate for a variety of downstream tasks in which the recovered multi-scale characteristics can be used for additional processing.
Researchers at NVIDIA have suggested Swin UNEt TRansformers (Swin UNETR). This unique architecture employs a U-shaped network with a Swin transformer as the encoder and connects it to a CNN-based decoder at various resolutions via skip connections. In the 2021 edition of the Multi-modal Brain Tumor Segmentation Challenge, the team demonstrated the effectiveness of their approach for the task of multi-modal 3D brain tumor segmentation (BraTS). In the validation phase, the model was one of the top-ranking approaches, and in the testing phase, it performed well.
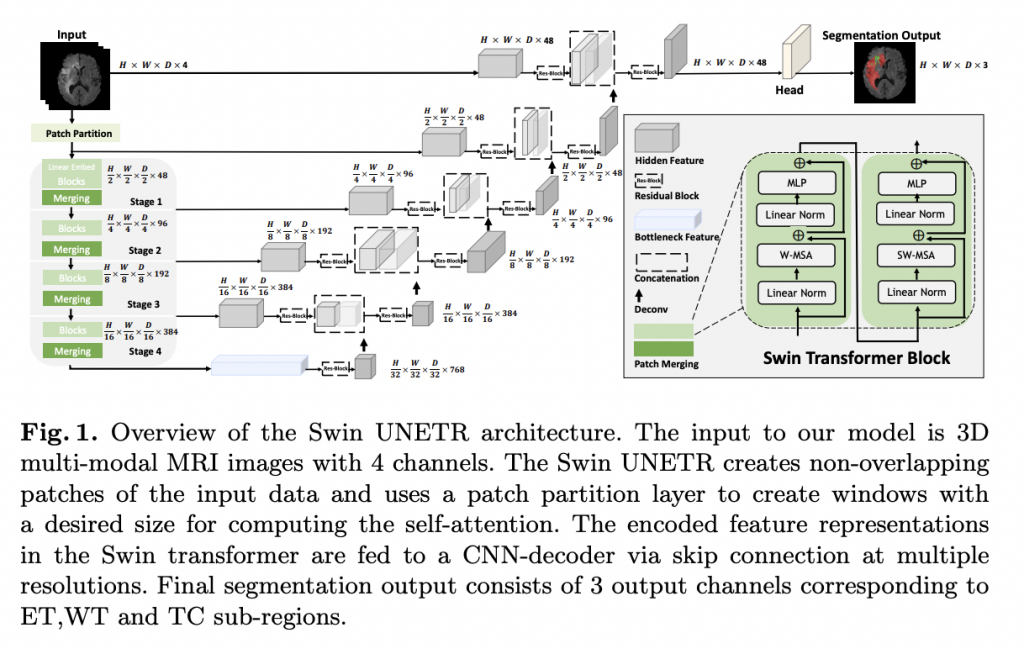
The encoder’s retrieved feature representations are employed in the decoder via skip links at each resolution in Swin UNETR’s U-shaped network design. Following that, a deconvolutional layer is used to improve the resolution of the feature maps by a factor of two, and the outputs are concatenated with the previous stage’s outputs.
Swin UNETR is trained on a DGX-1 cluster with 8 NVIDIA V100 GPUs using PyTorch3 and MONAI4. According to non-zero voxels, the team normalizes all input images to have a zero mean and unit standard deviation. During training, random patches of 128 128 128 were chopped from 3D image volumes. For all three axes, they apply a random axis mirror flip with a chance of 0.5.
In addition, the researchers used random per channel intensity shift, and random scale of intensity data augmentation transforms on the input image channels. Per GPU, the batch size was set to one. With a linear warmup and a cosine annealing learning rate scheduler, all models were trained for a total of 800 epochs.
By offering a 3D MRI dataset with voxel-wise ground truth labels annotated by clinicians, the BraTS challenge intends to evaluate state-of-the-art algorithms for the semantic segmentation of brain tumors. Data was gathered from a variety of universities utilizing various MRI scanners. Three tumor sub-regions are annotated: the enhancing tumor, peritumoral edema, and necrotic and non-enhancing tumor core. Whole Tumor (WT), Tumor Core (TC), and Enhancing Tumor (ET) were the three nested sub-regions created from the annotations (ET).
Researchers compared Swin UNETR’s performance in the internal cross-validation split to that of earlier winning methodologies such as SegResNet, nnU-Net, and TransBTS. The latter is a ViT-based technique that is specifically designed for brain tumor semantic segmentation. In all five-folds and on average for all semantic classes, the suggested Swin UNETR model surpasses all competing techniques (e.g., ET, WT, TC). Swin UNETR outperforms the closest rival techniques by 0.7 percent, 0.6 percent, and 0.4 percent in the ET, WT, and TC classes and by 0.5 percent on average across all classes in all folds.
Swin UNETR’s excellent success in brain tumor segmentation compared to other top-performing models is mainly due to its capacity to learn multi-scale contextual information in its hierarchical encoder via self-attention modules and effective modeling of long-range dependencies.
Conclusion
Swin UNETR, a revolutionary architecture for semantic segmentation of brain tumors utilizing multi-modal MRI data, was introduced by NVIDIA researchers. The suggested architecture has a U-shaped network design, with a Swin transformer serving as the encoder and a CNN-based decoder coupled to the encoder via skip links at various resolutions. In the validation phase, the model ranks among the top-performing techniques, and in the testing phase, it outperforms the competition. Swin UNETR, according to the researchers, might serve as the foundation for a new class of transformer-based models with hierarchical encoders for brain tumor segmentation.
Paper: https://arxiv.org/pdf/2201.01266v1.pdf
Github: https://github.com/Project-MONAI/research-contributions/tree/master/SwinUNETR
Suggested

Credit: Source link


Comments are closed.